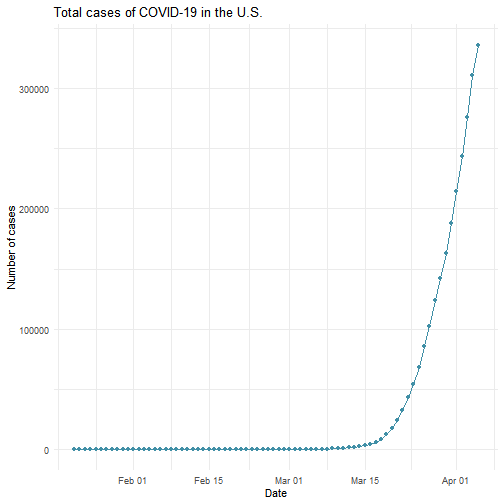
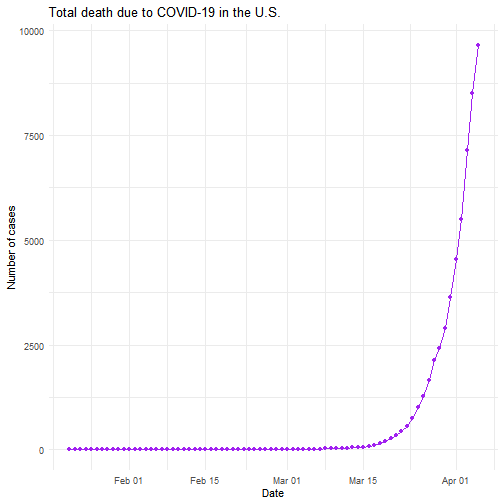
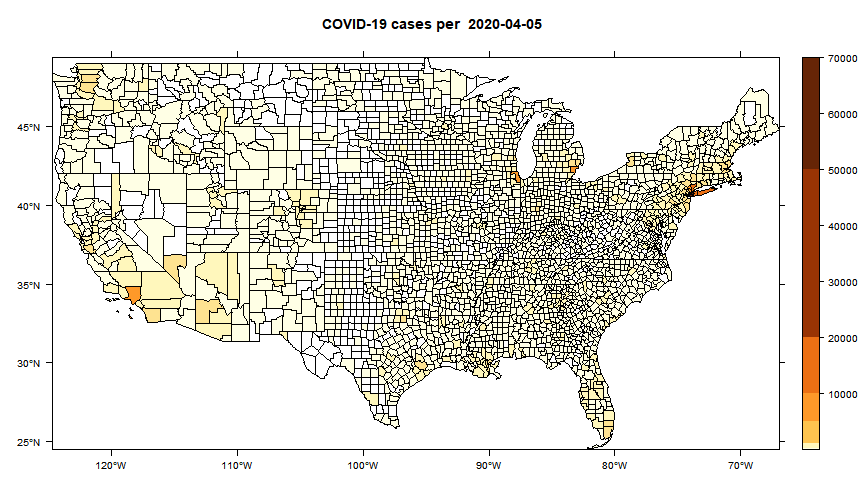
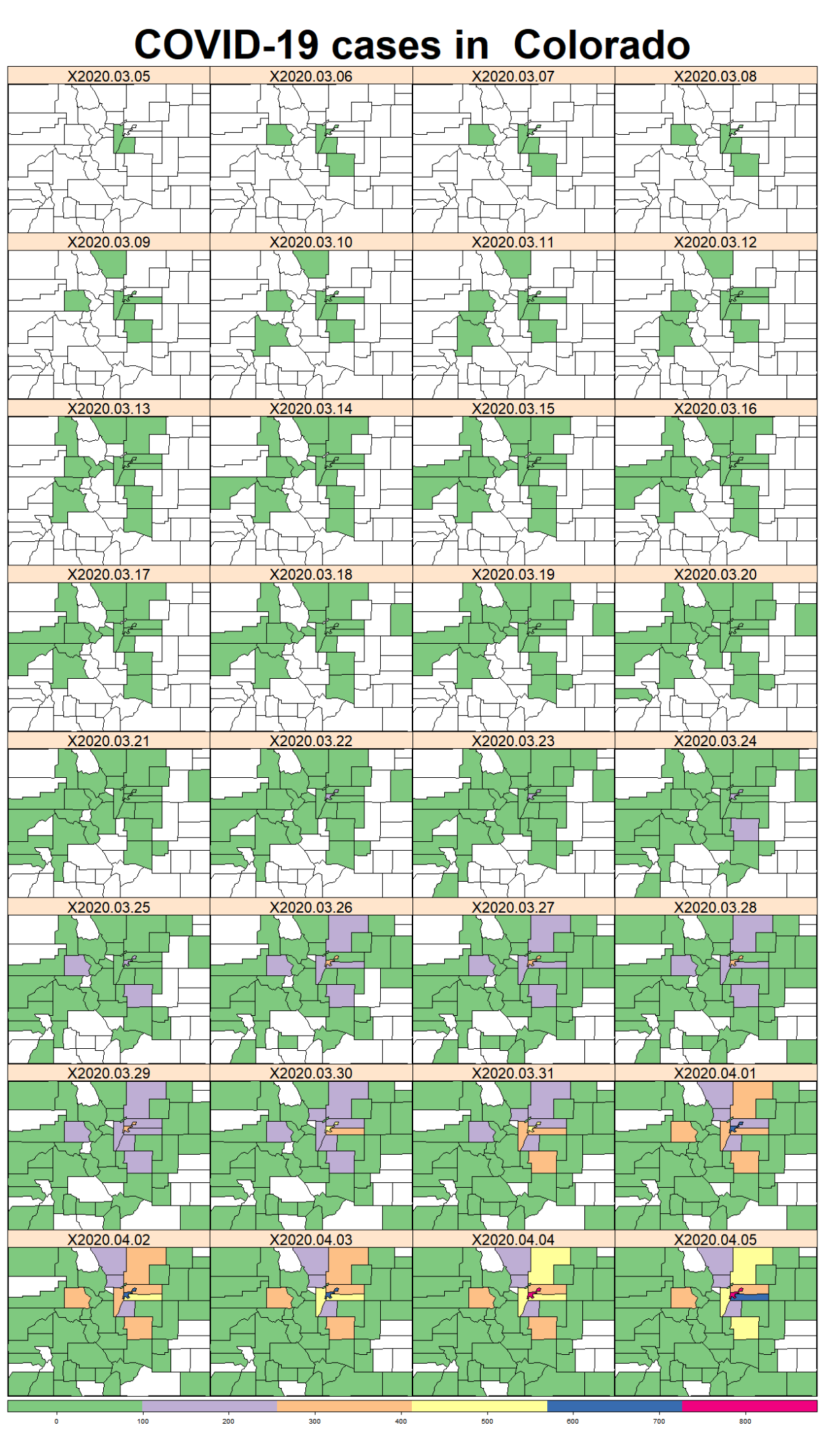
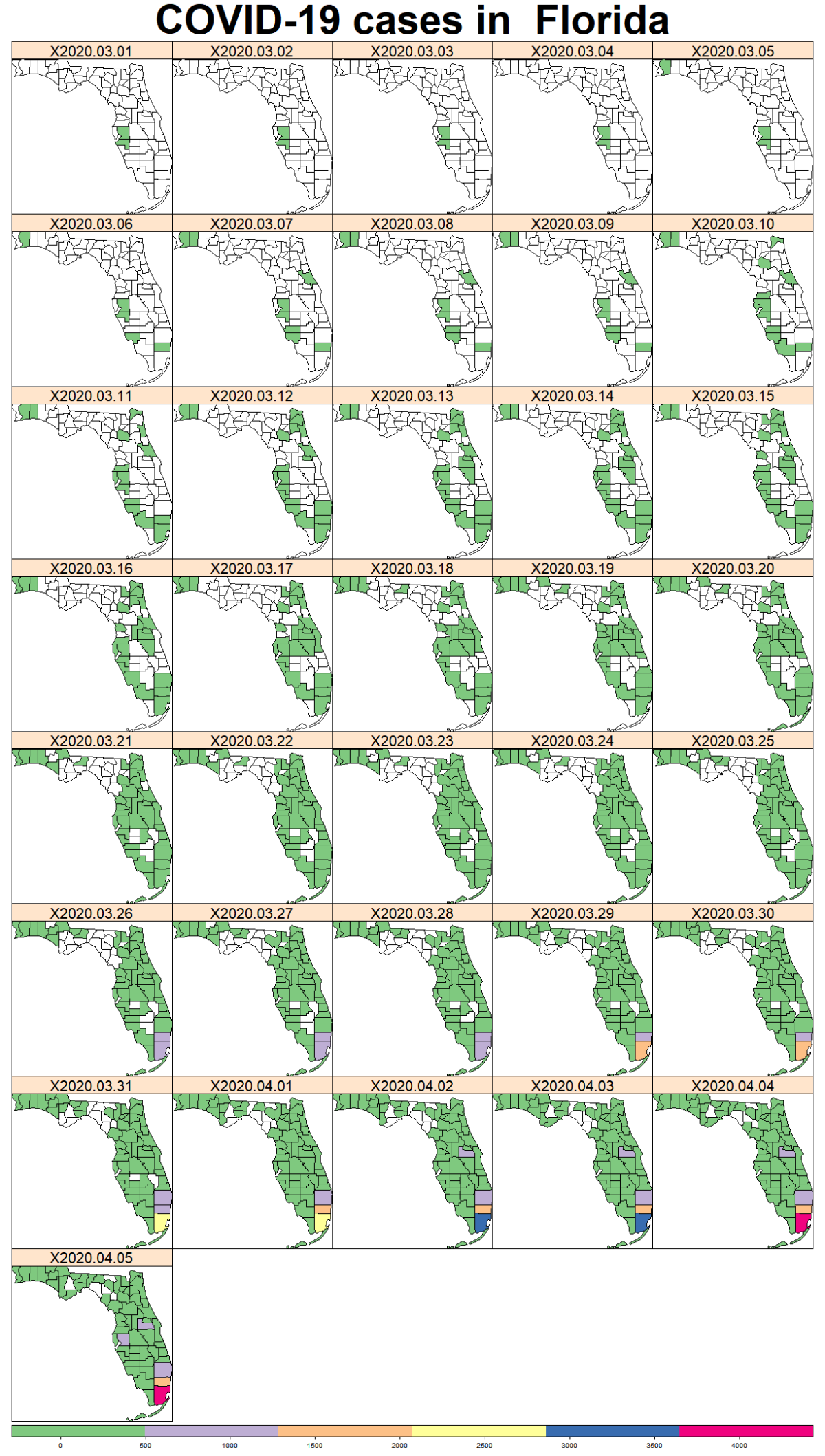
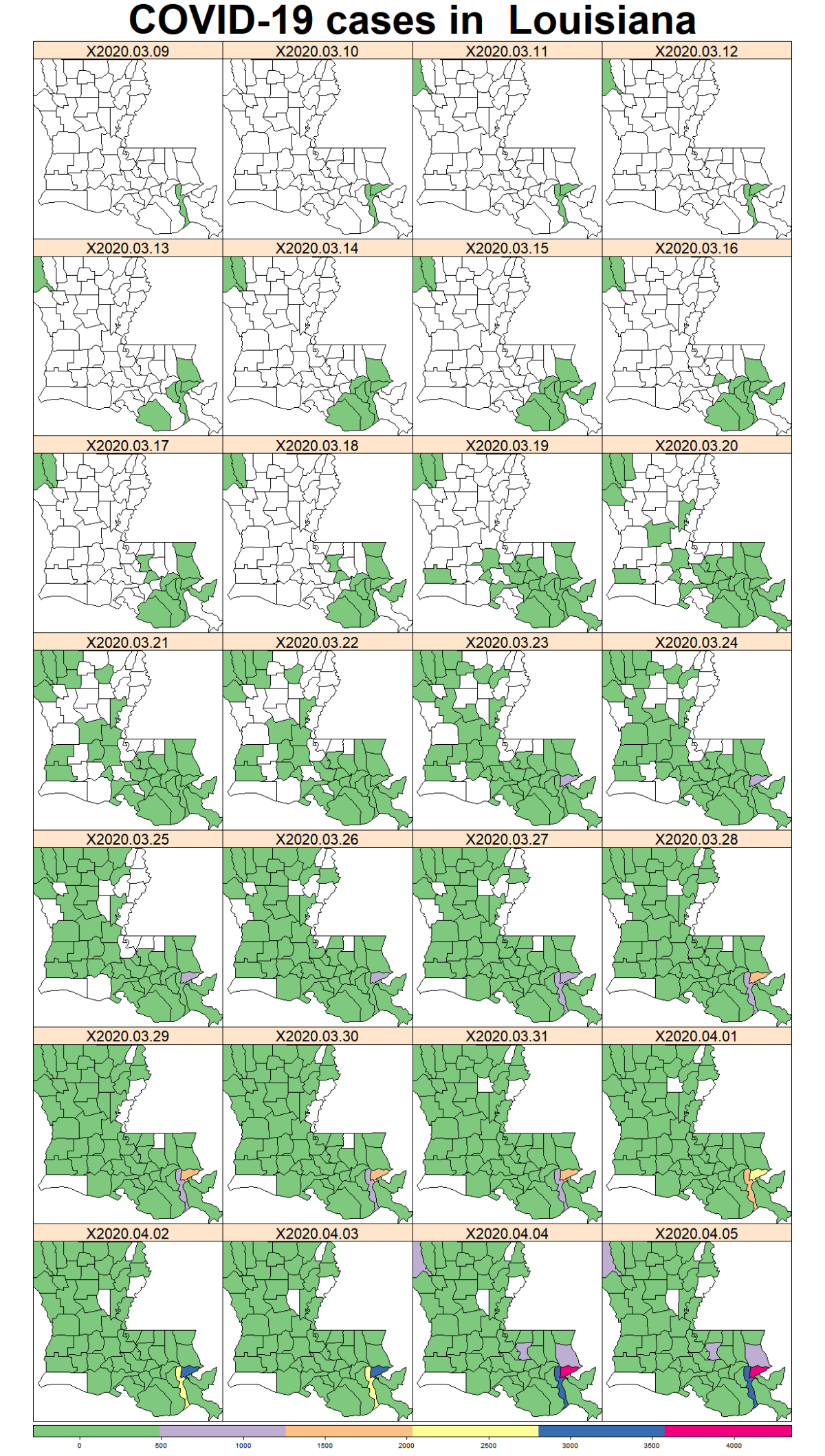
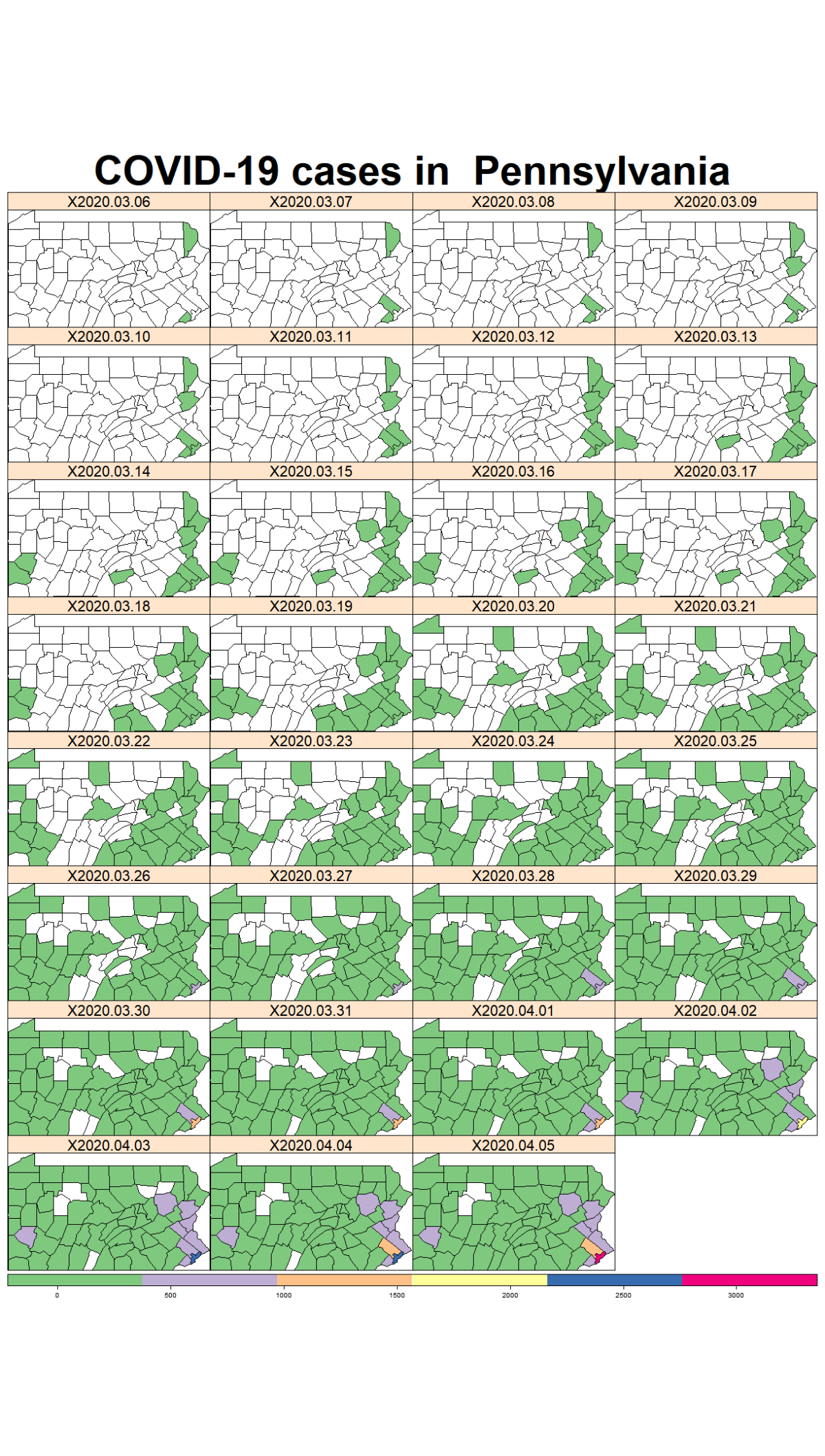
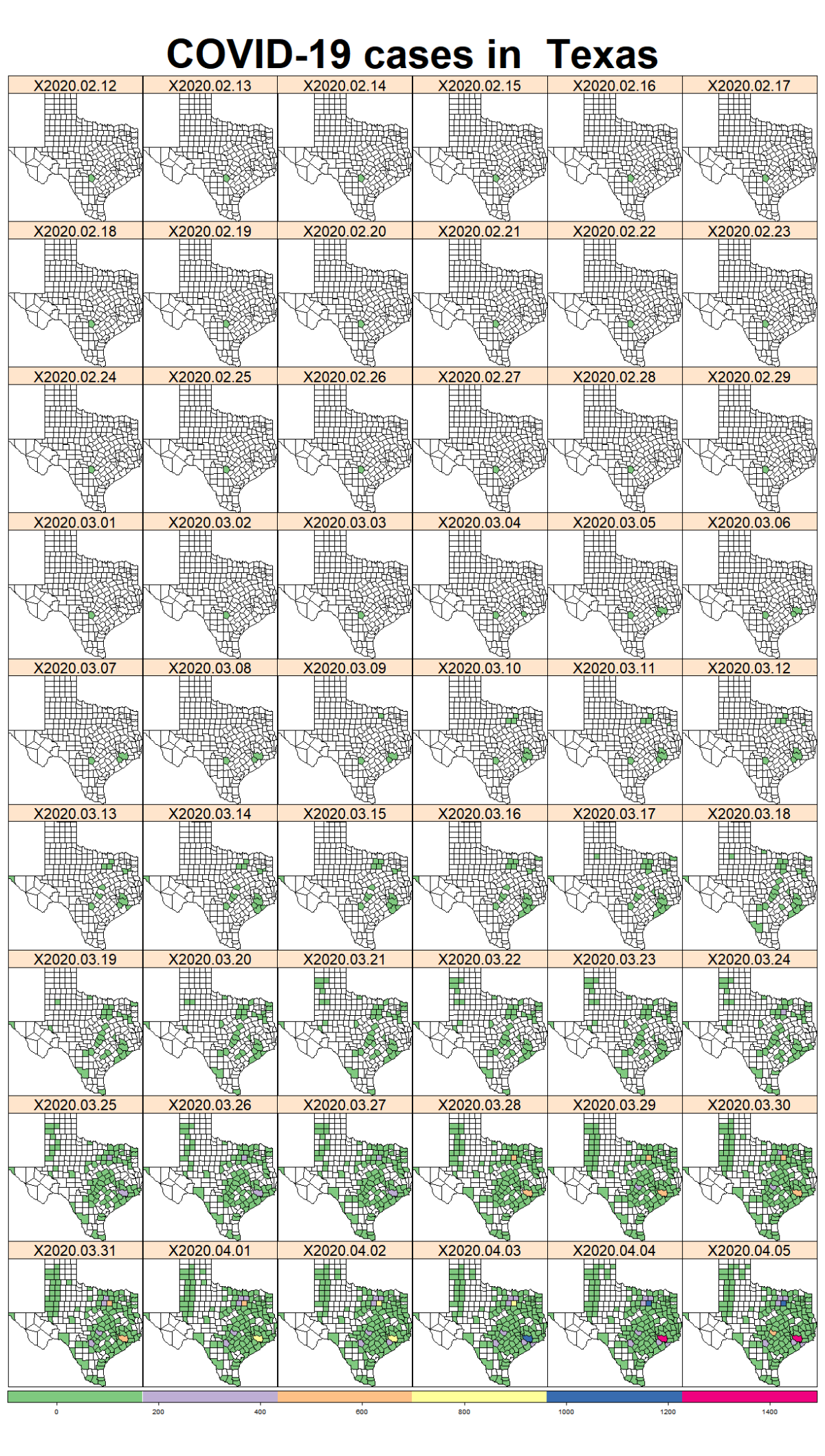
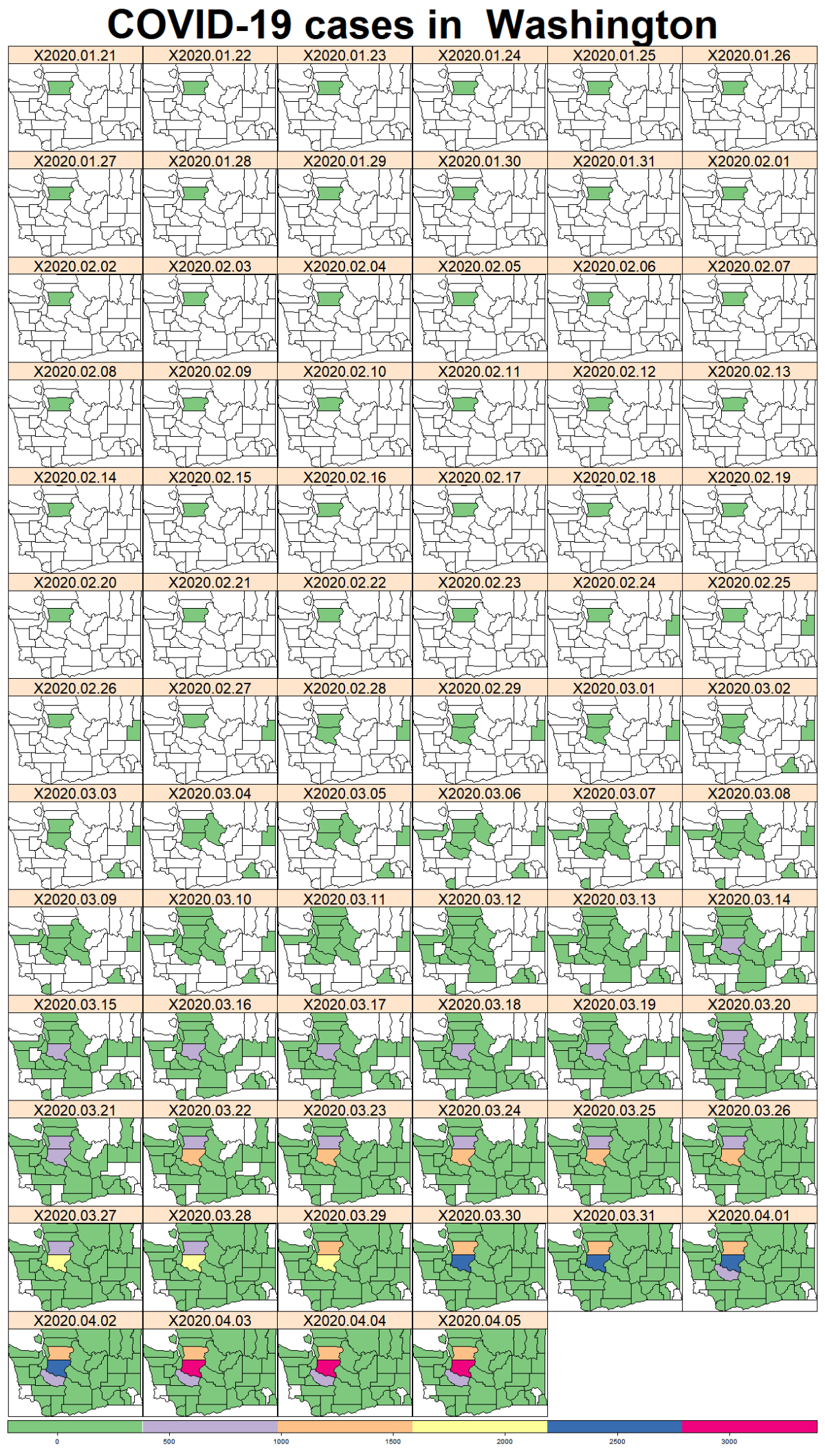
While the majority of states in the U.S. provide COVID-19 county level data, Pennsylvania and Illinois are among few states, as far as I know, that provide COVID-19 data in both county level and zip code level. Pennsylvania provides the number of positive and negative cases per zip code. Illinois provides not only the number of positive and negative cases but also the demographics information (e.g., age composition of positive case, race, and gender distribution) per zip code.
Illinois has more elaborate COVID-19 data. Hence, there are more things to explore with this data. From Illinois data, I want to focus on doing some analysis using zip code level data of Cook County, the most populous county in Illinois. As a former South Side Chicago and Chicago West suburb resident, where both are located in Cook County, I want to know how much COVID-19 impacts Chicago area communities, where I resided for almost 7 years.
My objective is to establish age-adjusted rate of COVID-19 cases by zip code in Cook County.
I am using R to calculate the age-adjusted rate and to visualize the distribution of COVID-19 rate. I want to visualize the distribution of the rates geographically. This allows me to identify which areas are hit harder by COVID-19 from the map.
I collect the data from various sources to do the analysis:
This data in this website is available in JSON format. I extract the data with Python script. I will publish the script anytime in the future.
This shapefile consists of the boundaries of block groups. Since I need the zip code boundaries, I dissolved multiple polygons of block groups with a common zip code into one new zip code polygon. I did this process in QGIS. I know it sounds like too much work to go back and forth between QGIS and R. But, I felt it was easier and faster to do it this way rather than doing it in R.
The ACS data that I downloaded provides and summarizes the data of population for every single census tract in Cook County. It consists of demographic, social, economic, and housing data.
Since I want to calculate the age-adjusted rate, I download the number of population for each age group.
The analysis will be per zip code level, but the number of population from ACS data is available per census tract level. I need to assign each census tract to zip codes. To do this, I use a crosswalk table which makes it possible to allocate census tract to zip codes. So, I join the ACS table with the tract-zip crosswalk table to extract the number of population from ACS table per zip code.
The projected 2010 U.S. population is used as the standard population, with eight groups aged 0 to 19, 20 to 29, 30 to 39, 40 to 49, 50 to 59, 60 to 69, 70 to 79, and 80 years or older.
The R code to work on this analysis is available below.
#------------#
# Libraries #
#------------#
library(raster)
library(sf)
library(dplyr)
library(ggplot2)
library(RColorBrewer)
library(rgdal)
library(scales)
library(tmap)
library(readxl)
library(ggpubr)
#-------------------#
# Reading the data #
#-------------------#
## Read Cook County zip code shapefile
cook_zip <- readOGR('.../cook_zip_edit.shp')
cook_zipSF <- st_as_sf(cook_zip)
# qtm(cook_zipSF, fill=NA)
## Read Illinois COVID-19 case per zip code
il_covid <- read.csv(file = '.../covid19_il_2020-05-07.csv')
il_covid$zipcode <- as.factor(il_covid$zip)
## Read Cook County ACS 5 years estimate from 2018
cook_acs <- read_excel('.../ACS5Y2018_IL.xlsx', sheet = 'Sheet1')
cook_acs$tract_new <- as.factor(trim(cook_acs$tract))
## Read Cook County crosswalk table of zip code and tract
crosswalk <- read.csv('.../TRACT_ZIP_122018.csv')
crosswalk$tract_new <- as.factor(crosswalk$tract)
#----------------#
# Joining tables #
#----------------#
## Join shapefile and Cook County IL covid data
cook_covid_zip <- inner_join(cook_zipSF, il_covid,
by = c('ZCTA5CE10' = 'zipcode'))
## alocate the tract to zip code
acs_tract <- inner_join(cook_acs, crosswalk[
c('tract_new', 'zip', 'res_ratio', 'tract')],
by = 'tract_new')
### adjusting the number of population with the res_ratio
acs_tract <- acs_tract %>%
mutate_at(
c('TotPop', 'TotPopMale', 'TotPop0to19', 'TotPop20to29',
'TotPop30to39', 'TotPop40to49', 'TotPop50to59',
'TotPop60to69', 'TotPop70to79', 'TotPopHE80',
'TotHH', 'Pop25', 'Pop25Bachelor', 'TotPopForeign',
'Pop5', 'Pop5Eng', 'Pop5OtherLang', 'PopLabor',
'PopLaborUnempl', 'Work16', 'Work16Drive',
'Work16Carpool', 'Work16Public', 'Work16Walk',
'Work16Other', 'Work16WFH', 'TotHHSNAP', 'PopCivil',
'PopCivilInsurance', 'TotHouse', 'TotHouseVacant',
'TotPopWhite', 'TotPopBlack', 'TotPopAsian',
'TotPopOther', 'TotPopHisp'),
funs(Aj = .*res_ratio)
)
acs_zip <- acs_tract %>%
group_by(zip) %>%
summarise_at(c('TotPop_Aj', 'TotPopMale_Aj',
'TotPop0to19_Aj', 'TotPop20to29_Aj',
'TotPop30to39_Aj', 'TotPop40to49_Aj',
'TotPop50to59_Aj', 'TotPop60to69_Aj',
'TotPop70to79_Aj', 'TotPopHE80_Aj',
'TotHH_Aj', 'Pop25_Aj', 'Pop25Bachelor_Aj',
'TotPopForeign_Aj', 'Pop5_Aj', 'Pop5Eng_Aj',
'Pop5OtherLang_Aj', 'PopLabor_Aj',
'PopLaborUnempl_Aj', 'Work16_Aj',
'Work16Drive_Aj', 'Work16Carpool_Aj',
'Work16Public_Aj','Work16Walk_Aj',
'Work16Other_Aj', 'Work16WFH_Aj',
'TotHHSNAP_Aj', 'PopCivil_Aj',
'PopCivilInsurance_Aj','TotHouse_Aj',
'TotHouseVacant_Aj', 'TotPopWhite_Aj',
'TotPopBlack_Aj', 'TotPopAsian_Aj',
'TotPopOther_Aj', 'TotPopHisp_Aj'), sum)
acs_zip$zipcode <- as.factor(acs_zip$zip)
## join acs_zip and covid-19 cases
acs_zip_covid <- inner_join(acs_zip, il_covid, by =c('zipcode'= 'zipcode'))
#--------------------------------------------#
# Create age-adjusted rate of COVID-19 cases #
#--------------------------------------------#
### Age-adjusted rates take into account the age-distribution of a population.
### Age-adjusted rates are calculated by dividing the number of positive COVID-19 cases
### by the ACS estimated population in that zip code and age group,
### then age-adjusting to the 2010 U.S. Census, and multiplying by 10,000.
### age-adjusted 0-19 yr
acs_zip_covid$crude_0to19 <- (acs_zip_covid$ageLT20/acs_zip_covid$TotPop0to19_Aj)*10000
acs_zip_covid$crude_weight_0to19 <- (acs_zip_covid$ageLT20/acs_zip_covid$TotPop0to19_Aj)*
((20201362+20348657+20677194+22040343)/308745538)*10000
### age-adjusted 20-29 yr
acs_zip_covid$crude_20to29 <- (acs_zip_covid$age20_29/acs_zip_covid$TotPop20to29_Aj)*10000
acs_zip_covid$crude_weight_20to29 <- (acs_zip_covid$age20_29/acs_zip_covid$TotPop20to29_Aj)*
((21585999+21101849)/308745538)*10000
### age-adjusted 30-39 yr
acs_zip_covid$crude_30to39 <- (acs_zip_covid$age30_39/acs_zip_covid$TotPop30to39_Aj)*10000
acs_zip_covid$crude_weight_30to39 <- (acs_zip_covid$age30_39/acs_zip_covid$TotPop30to39_Aj)*
((19962099+20179642)/308745538)*10000
### age-adjusted 40-49 yr
acs_zip_covid$crude_40to49 <- (acs_zip_covid$age40_49/acs_zip_covid$TotPop40to49_Aj)*10000
acs_zip_covid$crude_weight_40to49 <- (acs_zip_covid$age40_49/acs_zip_covid$TotPop40to49_Aj)*
((20890964+22708591)/308745538)*10000
### age-adjusted 50-59 yr
acs_zip_covid$crude_50to59 <- (acs_zip_covid$age50_59/acs_zip_covid$TotPop50to59_Aj)*10000
acs_zip_covid$crude_weight_50to59 <- (acs_zip_covid$age50_59/acs_zip_covid$TotPop50to59_Aj)*
((22298125+19664805)/308745538)*10000
### age-adjusted 60-69 yr
acs_zip_covid$crude_60to69 <- (acs_zip_covid$age60_69/acs_zip_covid$TotPop60to69_Aj)*10000
acs_zip_covid$crude_weight_60to69 <- (acs_zip_covid$age60_69/acs_zip_covid$TotPop60to69_Aj)*
((16817924+12435263 )/308745538)*10000
### age-adjusted 70-79 yr
acs_zip_covid$crude_70to79 <- (acs_zip_covid$age70_79/acs_zip_covid$TotPop70to79_Aj)*10000
acs_zip_covid$crude_weight_70to79 <- (acs_zip_covid$age70_79/acs_zip_covid$TotPop70to79_Aj)*
((9278166+7317795 )/308745538)*10000
### age-adjusted 80+ yr
acs_zip_covid$crude_HE80 <- (acs_zip_covid$ageHT80/acs_zip_covid$TotPopHE80_Aj)*10000
acs_zip_covid$crude_weight_HE80 <- (acs_zip_covid$ageHT80/acs_zip_covid$TotPopHE80_Aj)*
((5743327+3620459+1448366+371244+53364)/308745538)*10000
### Replacing NaN with 0 so crude and age adjusted rate column do not contain NaN.
### (This happens because the number of cases in certain age = 0
### and number of population of certain age = 0)
acs_zip_covid$crude_HE80[is.nan(acs_zip_covid$crude_HE80)] <- 0
acs_zip_covid$crude_weight_HE80[is.nan(acs_zip_covid$crude_weight_HE80)] <- 0
### Calulate the crude rate
acs_zip_covid$crude_rate <- acs_zip_covid$crude_0to19 +
acs_zip_covid$crude_20to29 + acs_zip_covid$crude_30to39 +
acs_zip_covid$crude_40to49 + acs_zip_covid$crude_50to59 +
acs_zip_covid$crude_60to69 + acs_zip_covid$crude_70to79 +
acs_zip_covid$crude_HE80
### Calculate the Age-adjusted rate of COVID-19
acs_zip_covid$age_adjusted_rate <- acs_zip_covid$crude_weight_0to19 +
acs_zip_covid$crude_weight_20to29 + acs_zip_covid$crude_weight_30to39 +
acs_zip_covid$crude_weight_40to49 + acs_zip_covid$crude_weight_50to59 +
acs_zip_covid$crude_weight_60to69 + acs_zip_covid$crude_weight_70to79 +
acs_zip_covid$crude_weight_HE80
### removing non Cook County IL zip code
acs_zip_covid <- acs_zip_covid %>% filter(
!zipcode %in% c('60102', '60448', '60491',
'60417', '60423', '60449',
'60523', '60126', '60484',
'60015', '60521', '60527',
'60172', '60118'))
## join shapefile and cases table
zip_case <- inner_join(cook_zipSF, acs_zip_covid, by = c('ZCTA5CE10' = 'zipcode'))
#------------------------#
# Descriptive Statistics #
#------------------------#
## Age distribution of confirmed COVID-19 cases and the population
### Reshaping data from wide to long format
zip_caseSP <- as(zip_case, 'Spatial')
zip_caseSP_df <- zip_caseSP@data
selected_col_df <- zip_caseSP_df[c('zip.x', 'ageLT20', 'age20_29',
'age30_39', 'age40_49',
'age50_59', 'age60_69',
'age70_79', 'ageHT80',
'TotPop0to19_Aj', 'TotPop20to29_Aj',
'TotPop30to39_Aj', 'TotPop40to49_Aj',
'TotPop50to59_Aj', 'TotPop60to69_Aj',
'TotPop70to79_Aj', 'TotPopHE80_Aj',
'white', 'black', 'hispanic',
'asian', 'other', 'NH_PI', 'AI_AN',
'TotPopWhite_Aj', 'TotPopBlack_Aj',
'TotPopHisp_Aj', 'TotPopAsian_Aj',
'TotPopOther_Aj')]
selected_col_df$others <- selected_col_df$other + selected_col_df$NH_PI +
selected_col_df$AI_AN
agegroup_df <- reshape(data = selected_col_df,
idvar = 'zip.x',
varying = list(numcase=c('ageLT20', 'age20_29',
'age30_39', 'age40_49',
'age50_59', 'age60_69',
'age70_79', 'ageHT80'),
numpop=c('TotPop0to19_Aj', 'TotPop20to29_Aj',
'TotPop30to39_Aj', 'TotPop40to49_Aj',
'TotPop50to59_Aj', 'TotPop60to69_Aj',
'TotPop70to79_Aj', 'TotPopHE80_Aj')),
direction="long",
v.names = c("numcase","numpop"),
times=c('0 to 19', '20 to 29', '30 to 39', '40 to 49',
'50 to 59', '60 to 69', '70 to 79', '80+'),
sep="")
age_summ <- agegroup_df %>% group_by(time) %>%
summarise(case = sum(numcase), pop=sum(numpop))
age_summ$pctcase <- age_summ$case*100/(sum(age_summ$case))
age_summ$pctpop <- age_summ$pop*100/sum(age_summ$pop)
p1 <- ggplot(age_summ, aes(x=time, y=pctcase)) +
geom_bar(stat="identity", fill="thistle2")+
xlab('Age group') + ylab('%') +
theme_minimal() +
ggtitle('Fig 1. Age distribution of COVID-19 cases in Cook County, \nas of May 7, 2020 (%)')
p2 <- ggplot(age_summ, aes(x=time, y=pctpop)) +
geom_bar(stat="identity", fill="seagreen3")+
xlab('Age group') + ylab('%') +
theme_minimal() +
ggtitle('Fig 2. Age distribution of Cook County population (%)')
ggarrange(p1, p2,
ncol = 1, nrow = 2)
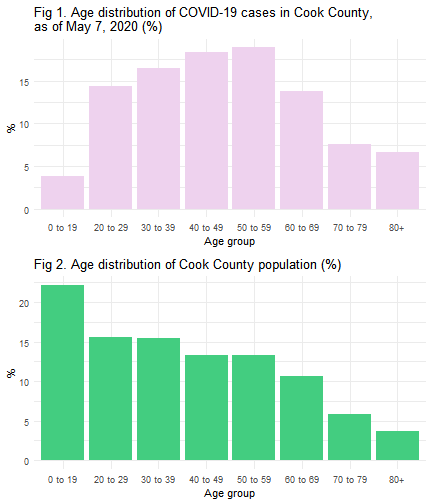
Until May 7, 2020, a total of 47,480 (including 10 cases of unknown age) confirmed cases of COVID-19 occurred in Cook County. As shown in Figure 1, around 18.9% of COVID-19 confirmed cases in Cook County were in their 50s, followed by people in their 40s (18.3%), 30s (16.4%), and 20s (14.3%).
## Race distribution of confirmed COVID-19 cases and the population
race_df <- reshape(data = selected_col_df,
idvar = 'zip.x',
varying = list(numcase=c('white', 'black', 'hispanic',
'asian', 'others'),
numpop=c('TotPopWhite_Aj', 'TotPopBlack_Aj',
'TotPopHisp_Aj', 'TotPopAsian_Aj',
'TotPopOther_Aj')),
direction="long",
v.names = c("numcase","numpop"),
times=c('White', 'Black', 'Hispanic', 'Asian', 'Others'),
sep="")
race_summ <- race_df %>% group_by(time) %>% summarise(case = sum(numcase), pop=sum(numpop))
race_summ$pctcase <- race_summ$case*100/(sum(race_summ$case))
race_summ$pctpop <- race_summ$pop*100/sum(race_summ$pop)
p3 <- ggplot(race_summ, aes(x=time, y=pctcase)) +
geom_bar(stat="identity", fill="bisque2")+
xlab('Race') + ylab('%') +
theme_minimal() +
ggtitle('Fig 3. Race distribution of COVID-19 cases in Cook County \nas of May 7, 2020 (%)')
p4 <- ggplot(race_summ, aes(x=time, y=pctpop)) +
geom_bar(stat="identity", fill="seagreen3")+
xlab('Race') + ylab('%') +
theme_minimal() +
ggtitle('Fig 4. Race distribution of Cook County population (%)')
ggarrange(p3, p4,
ncol = 1, nrow = 2)
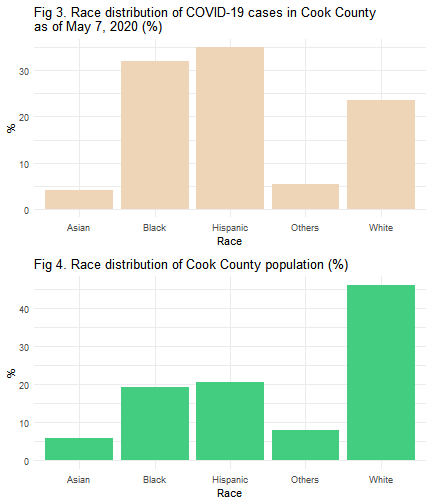
In Figure 4, even though Hispanics make up 20.6% of the Cook County population, they account for 35% of cases and surpass all other race groups in the total number of COVID-19 cases in Cook County, as shown in Figure 3. Meanwhile, black residents make up almost 32% of COVID-19 cases, followed by white residents (23.4%).
There are multiple factors that contribute to the variation of COVID-19 infections among different age group and races. Living arrangement such as larger household size with multiple generations living under the same roof among Hispanic families1,2 may make it more likely that Hispanics are tested positive.
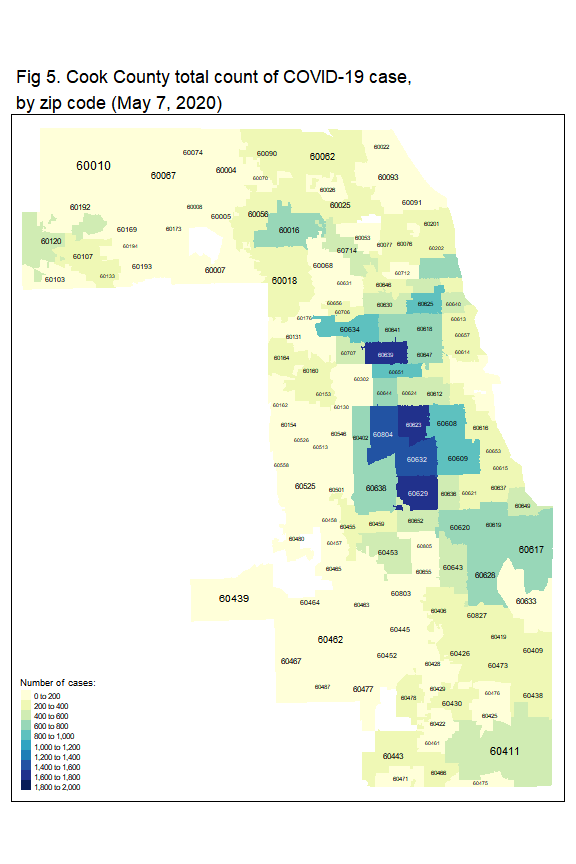
Figure 5 shows the number of cases per zip code across Cook County per May 7, 2020. The lowest and the highest number of cases are 0 and 1,762, respectively. The median number of cases is 178. Some community areas with higher number of cases located in Northwest Side of Chicago, West Side, Southwest Side of Chicago, South Side, and Cicero. The majority of residents in these communities are Hispanic or black.
### number of cases
tm_shape(zip_case) +
tm_fill('positive',
title = 'Number of cases:',
style = 'fixed',
breaks = seq(0,2000,200),
palette = 'YlGnBu')+
tm_layout(legend.title.size = 0.9,
main.title = 'Fig 5. Cook County total count of COVID-19 case,\nby zip code (May 7, 2020)',
title.size = 0.9) +
tm_text('ZCTA5CE10', size='AREA')
Almost all disease happen at different rates in different age groups. Some diseases happen more often among elderly and some happen more often among the younger people. To control the effect of differences in population age distributions, the age-adjusted rates3 are calculated. Using age-adjusted rates will make the different groups more comparable because the adjusting minimizes the bias of age.
I employ direct standardization for adjusting. The first step is to divide the number of COVID-19 cases by the ACS estimated population in that zip code. The result of this division is called crude rates. As I use eight age groups (from 0 to 19 to 80 years or older), there will be eight crude rates in total per zip code. Finally, the age-adjusted rate is calculated by multiplying each crude rate by the proportion of U.S. 2010 standard population within each age group, and then summing the products.
Figure 6 and Figure 7 show geographic distribution of COVID-19 crude rate and age-adjusted rate, respectively.
### crude rate
tm_shape(zip_case) +
tm_fill('crude_rate',
title = 'Crude rates per \n10,000 people:',
style = 'fixed',
breaks = seq(0,2000,200),
palette = 'BuGn')+
tm_layout(legend.title.size = 1.2,
main.title = 'Fig 6. Cook County COVID-19 crude \nrates, by zip code (May 7, 2020)',
title.size = 0.9) +
tm_text('ZCTA5CE10', size='AREA')
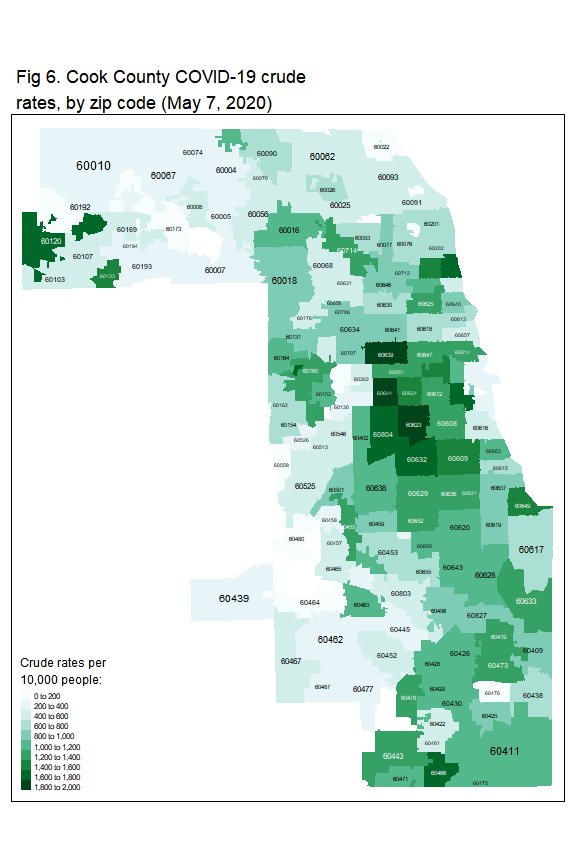
The crude rates range from 0 to 1864 per 10,000 people. After adjusting for age, the rates are now lower than the crude rates.
### age adjusted rate
tm_shape(zip_case) +
tm_fill('age_adjusted_rate',
title = 'Age adjusted rates \nper 10,000 people:',
style = 'fixed',
breaks = seq(0,250,25),
palette = 'Reds')+
tm_layout(legend.title.size = 1.2,
main.title = 'Fig 7. Cook County COVID-19 age adjusted rates,\nby zip code (May 7, 2020)',
title.size = 0.9) +
tm_text('ZCTA5CE10', size='AREA')
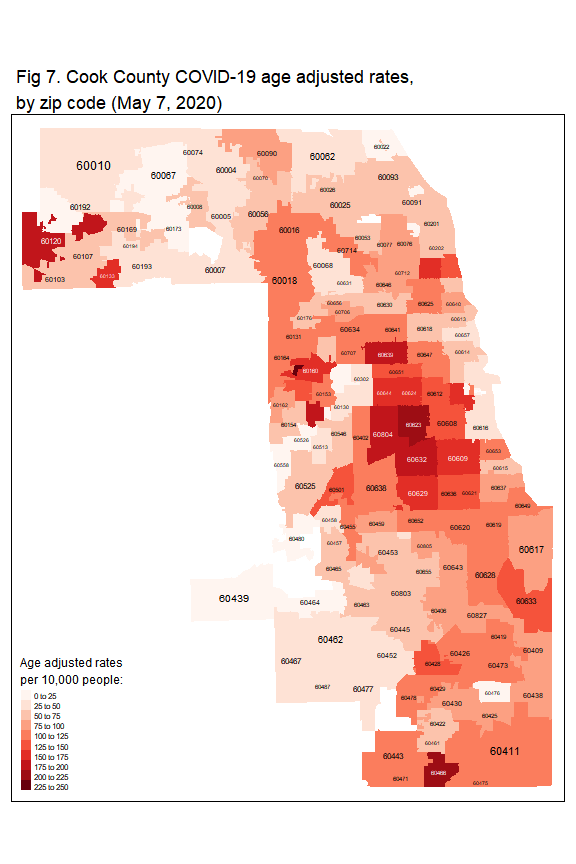
The age-adjusted rates range from 0 to 233 per 10,000 people. In Figure 7, the higher rates in Cook County are concentrated in:
- City of Chicago. In Chicago, these communities are hit harder by COVID-19: West Side (zip code: 60623, 60608, 60644, and 60624), Northwest side (zip code: 60639), Southwest side
(zip code: 60629), and South Side (zip code: 60609). - West part, i.e., Cicero (zip code: 60804), Stone Park (zip code: 60165), and Broadview (zip code: 60155)
- Northwest part, i.e., Elgin (zip code: 60120) and Hanover Park (zip code: 60133)
- South part, i.e., Park Forest (zip code: 60466)
References:
1. Cultural Insights: Communicating with Hispanics/Latinos https://www.cdc.gov/healthcommunication/pdf/audience/audienceinsight_culturalinsights.pdf
2. Illinois Latinos now have highest rate of coronavirus infections, IDPH data shows: https://abc7chicago.com/health/il-latinos-have-highest-rate-of-covid-19-infections-idph-data/6150864/
3. Calculating Age-adjusted Rates: https://seer.cancer.gov/seerstat/tutorials/aarates/definition.html